Next Generation Synthesis
Joseph Jacobson (MIT)
Recordings
- Will be linked after class
Presentations and slides
- Will be linked after class
Class assignments
Synthetic oligonucleotides (oligos) enable us to assemble novel DNA sequences or create copies of existing genes. This week's assignment will give you experience building genes and expressing them in bacteria.
Lab Task: Recode green fluorescent protein (GFP) to instead emit blue or cerulean light. Confirm success by transforming your assembled BFP and CFP variants into E coli cells.
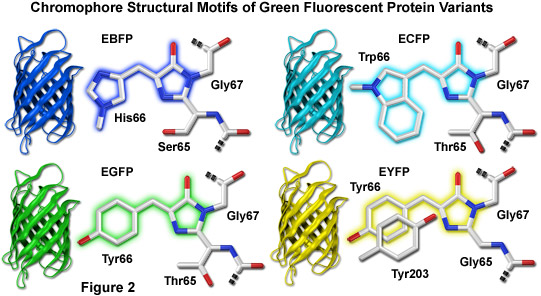
Theory Task: Design a gene. Describe a detailed workflow for constructing and expressing it. Identify how the parts of your genetic construct relate to DNA replication and the Central Dogma of Molecular Biology.
Hardware
| Function | Item |
|---|---|
| Polymerase Chain Reaction | Thermocycler |
| Incubating samples at 37C | Thermocycler, water bath, incubator, or body heat |
| Separating DNA by size | Gel electrophoresis box and power supply |
| Visualizing stained DNA | Blue-LED transilluminator |
| Preparing gel matrix and LB agar plates | Microwave and balance |
| DNA Extraction from TAE Agarose Gel | Microcentrifuge |
Software
| Function | Item |
|---|---|
| Mapping plasmids and recoding the GFP sequence | DNA sequence viewer, such as Benchling, Snapgene, Geneious, ApE, GenBeans, or text editor |
Wetware
| Function | Item |
|---|---|
| Amplifying DNA | DNA oligonucleotides and DNA polymerase mix to amplify GFP and pUC19 |
| Golden Gate Assembly | Type IIS restriction enzyme (i.e. BbsI-HF) and T7 DNA Ligase |
| Setting electrophoresis gel | Tris-acetate-EDTA (TAE) buffer and agarose |
| Setting agar plates | LB agar dehydrated culture media |
| Drug selection | Carbenicillin |
| Oligo Name | Sequence | Oligo Pair Partner | Pair Name |
|---|---|---|---|
| pUC19_GolGat_2026R | CACCACAGAAGACGATCAGCTCACTCAAAGGCGG | pUC19_GolGat_1F | pUC19_GolGat |
| pUC19_GolGat_1F | CACCACAGAAGACGACGGCATCCGCTTACAGACAA | pUC19_GolGat_2026R | pUC19_GolGat |
| pGreen_nGFP_GolGat_192R | CACCACAGAAGACGAAGTAGTGACTAGTGTTGGCCA | pGreen_nGFP_GolGat_1F | pGreen_nGFP |
| pGreen_nGFP_GolGat_1F | CACCACAGAAGACGATAACATGGCTAGCAAAGGAGAAGAACT | pGreen_nGFP_GolGat_192R | pGreen_nGFP |
| pGreen_cGFP_GolGat_575R | CACCACAGAAGACGAATAGGGCGAATTCGAGCTCG | pGreen_c(G/C/B)FP_GolGat_1F | pGreen_c(G/C/B)FP_GolGat |
| pGreen_cGFP_GolGat_1F | CACCACAGAAGACGATACTCTGTGCTATGGTGTTCAATGCT | pGreen_cGFP_GolGat_575R | pGreen_cGFP_GolGat |
| pGreen_cCFP_GolGat_1F | CACCACAGAAGACGATACTCTGACCTGGGGTGTTCAATGCT | pGreen_cGFP_GolGat_575R | pGreen_cCFP_GolGat |
| pGreen_cBFP_GolGat_1F | CACCACAGAAGACGATACTCTGACCCATGGTGTTCAATGCT | pGreen_cGFP_GolGat_575R | pGreen_cBFP_GolGat |
| T500_s1 | CTATGAGCAAAGCCCGCCGAAAGGCGGGCTTTTCTGTACA | T500_s2 | T500 |
| T500_s2 | GCCGTGTACAGAAAAGCCCGCCTTTCGGCGGGCTTTGCTC | T500_s1 | T500 |
| OR2OR1Pr_s1 | CTGATGAGCTAACACCGTGCGTGTTGACAATTTTACCTCTGGCGGTGATAATGGTTGCAG | OR2OR1Pr_s2 | OR2OR1Pr |
| OR2OR1Pr_s2 | CAAGCTGCAACCATTATCACCGCCAGAGGTAAAATTGTCAACACGCACGGTGTTAGCTCA | OR2OR1Pr_s1 | OR2OR1Pr |
| 5UTR_RBS_s1 | CTTGCAATAATTTTGTTTAACTTTAAGAAGGAGATA | 5UTR_RBS_s2 | 5UTR_RBS |
| 5UTR_RBS_s2 | GTTATATCTCCTTCTTAAAGTTAAACAAAATTATTG | 5UTR_RBS_s1 | 5UTR_RBS |
Brief Protocol
1 Resuspend oligos to make a 100 uM stock and then 10 uM dilutions that combine oligo pairs
- Read nmol amount of oligo from tube label
- For every nmol, add 10 ul water
- Vortex or flick tube and wait several minutes for the oligo to go into solution
2 In a separate tube combine 80 ul water and 10 ul of each 100 uM oligo in a pair to make 10 uM paired dilutions
- Oligo pairs that will be annealed as duplexes are named with the format X_s1 and X_s2. As an example, for X=T500:
| Item | Vol to Mix |
|---|---|
| T500_s1 | 10 ul |
| T500_s2 | 10 ul |
| Water | 80 ul |
- Oligo pairs that will be used together as PCR primers together are named with the format XGolgat_1F and X_Golgat##R or XY_Golgat_1F and X_Y'_Golgat##R.
3 Prepare Golden Gate annealed duplex fragments.
- In separate PCR tubes, mix the following
| Item | Vol to Mix |
|---|---|
| X_s1 + X_s2 dilution (10 uM) | 8 ul |
| T4 DNA ligase buffer (10x) | 1 ul |
| T4 Polynucleotide Kinase (T4 PNK) | 1 ul |
- Incubate mix at 37C for 1 hr
- Heat mixtures at 95C in thermocycler for 5 min and gradually cool to RT for 1 hr
4 Prepare the other Golden Gate fragments by PCR from plasmid X in primer pair X_Y_Golgat.
- In separate PCR tubes, mix the following 50 ul PCR reactions
| Item | Vol to Mix |
|---|---|
| X_Y_GolGat primer pair (10 uM) | 2.5 ul |
| Template plasmid X | 1 ul |
| dNTPS (10 uM) | 1 ul |
| Phusion polymerase buffer (5x) | 10 ul |
| Phusion DNA polymerase | 1 ul |
| Water | 34.5 ul |
- Run the following PCR program in your thermocycler with the lid set to 105 C
| Stage | Steps |
|---|---|
| 1 | Initial denaturation: 98C for 30 sec |
| 2 (30x cycles) | 1. Denaturation: 98C for 10 sec, 2. Annealing: 60C for 30 sec, 3. Extension: 72C for 30 sec/kb |
| 3 | Final extension: 72C for 5 min |
- Run finished PCR reaction on a 1% agarose TAE gel (see protocol homework 2)
- Purify the correct-sized band with a gel extraction kit (see kit manufacturer’s protocol)
5 Combine and assemble the Golden Gate fragments
- For each fragment from step 4 derived from pGreen_c(G/C/B)FP_GolGat_1F, mix the following in a thermocycler tube
| Item | Vol to Mix |
|---|---|
| 6 fragments from steps 3 and 4 | 1 ul each |
| Cutsmart buffer (10x) | 2 ul |
| ATP (10 mM) | 2 ul |
| T7 or T4 DNA ligase | 1 ul |
| BbsI-HF | 1 ul |
| Water | 8 ul |
- Run the following assembly program in your thermocycler with the lid set to 105 C
| Stage | Steps |
|---|---|
| 1 (30x cycles) | 1. Digest: 37C for 5 min, 2. Ligate: 16C for 5 min |
| 2 | Heat denature: 55C for 10 min |
6 Heat shock Golden Gate assembly into chemically competent E coli cells
- Thaw pre-aliquoted 25 ul E coli cells on ice for 30 min
- Add 2 ul of assembly to cells and keep on ice for 15 min
- Heat shock cells at 42C for 45 sec and immediately place on ice
- Mix 500 ul outgrowth media with cells and plate 100 ul