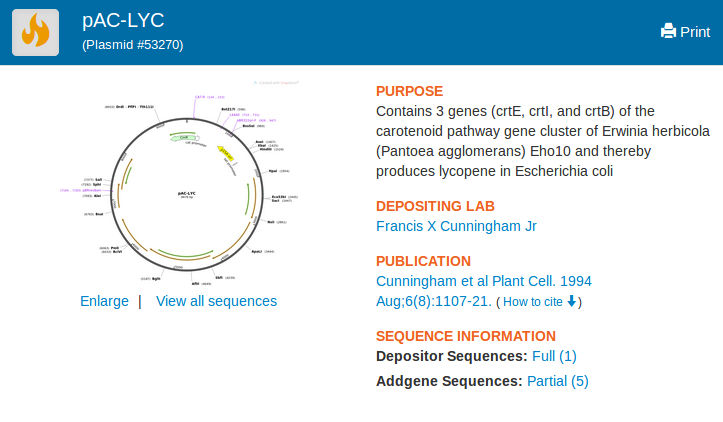
A. Lab Task: Test pAC-LYC plasmid
The pAC-LYC plasmid is available from Addgene, and contains three genes from Erwinia herbicola: CrtE, CrtI, and CrtB. This plasmid is provided by Addgene on an agar stab transformed into Top10 E. coli. That means that it can simply be streaked onto LB Agar with chloramphenicol to make lycopene-producing colonies that should be pink to red in color.
pAC-LYC plasmid can be found in addgene. The following is a screenshot of the addgene page
containing the plasmid.
A detailed view of the plasmid is shown below where Crt genes are clearly visible.
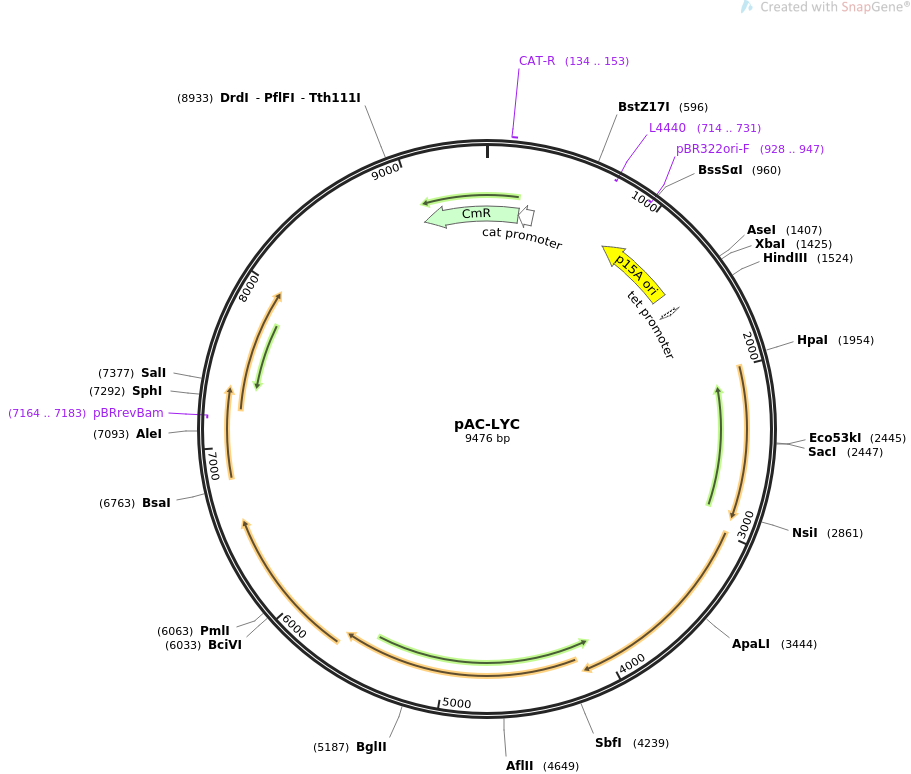
Ecoli with this plasmid can be obtained and streaked into agar media containing antibiotic
chloramphenicol to get red colored colonies.
B . Design strategies for increasing lycopene production
Using tools such as Biocyc (https://www.biocyc.org) and KEGG (http://www.kegg.jp), select enzymes that can be added to (or knocked out of) your lycopene-producing E. coli to increase the amount of lycopene they produce. The pAtipiTrc plasmid available in Addgene represents one possible strategy, as it over expresses a heterologous idi enzyme. Labs working with PCR and cloning may want to try replacing the idi gene in pAtipiTrc with alternative enzymes to see how they impact colony color.
The following metabolic pathway image show lycopene metabolism in plant hormones bio sysnthesys.
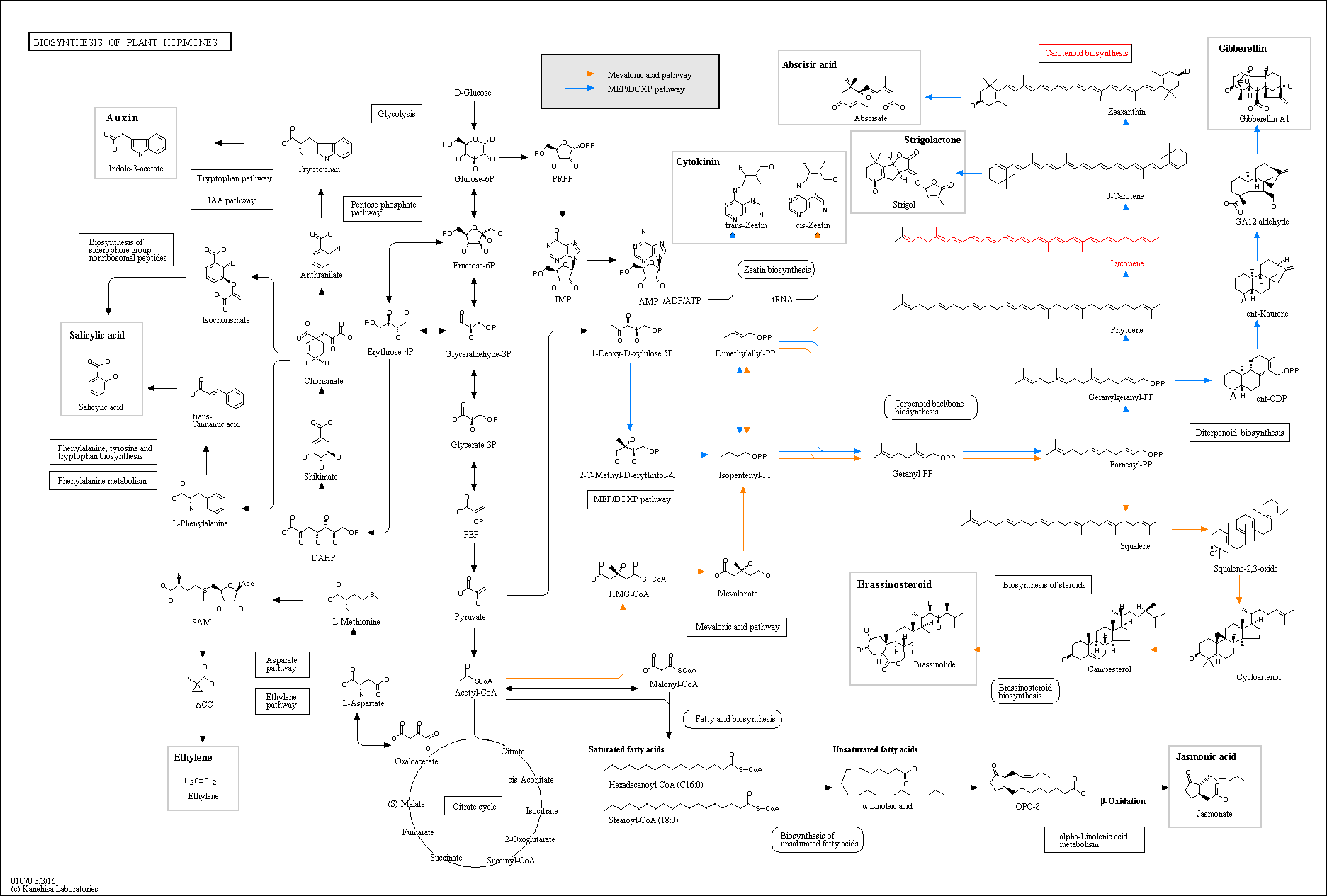
View full image
In the pathway you can see that Fernesyl-PP which is a precursor for lycopene getting converted to
Squaline. Also Geranylgeranyl-PP gets converted to ent-CDP. If we could knockout enzymes which catalyses
these reactions, then we can increase the production of lycopene. The following image shows the pathways to
be blocked.
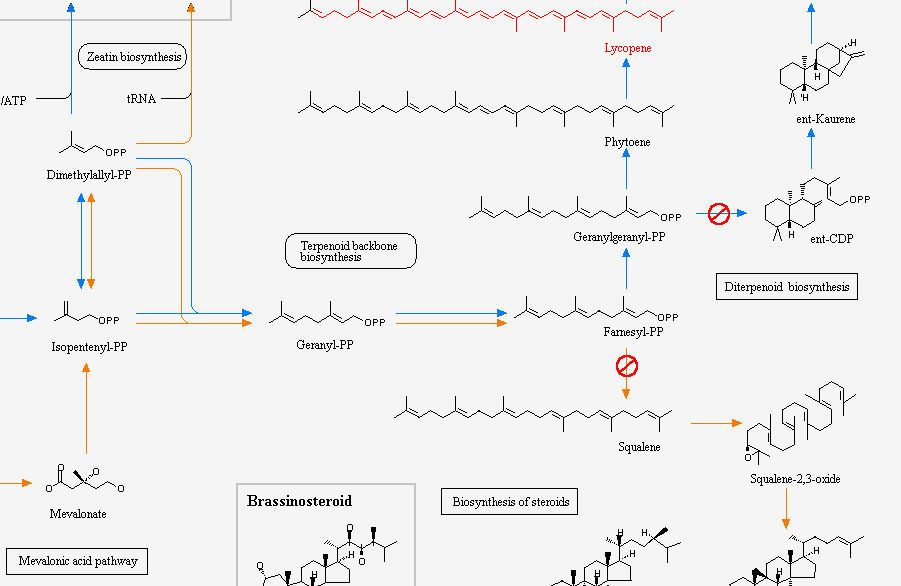
This is a part of carotenoid biosynthesys metabolic pathway showing lycopene synthesys.
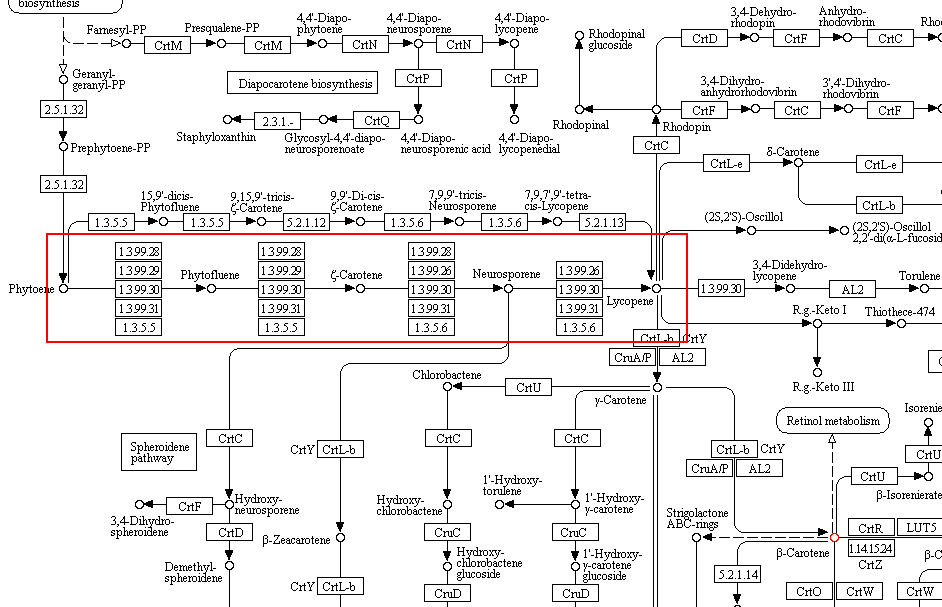
Over expression of any of the precursors of lycopene in this pathway also can increase its production.
C. Design strategies for converting lycopene to beta-carotene
Use Biocyc and/or KEGG to identify the enzyme that can convert the red pigment lycopene to the orange pigment beta-carotene. Plasmids that can be co-transformed with pAC-LYC that produce this enzyme can be found in Addgene. Labs working on miniprep and transformation can order these plasmids to observe switch the color of their E. coli from red to orange.
Carotene can be found in KEGG website in this link.
These are the details you can find about the compound in the website.
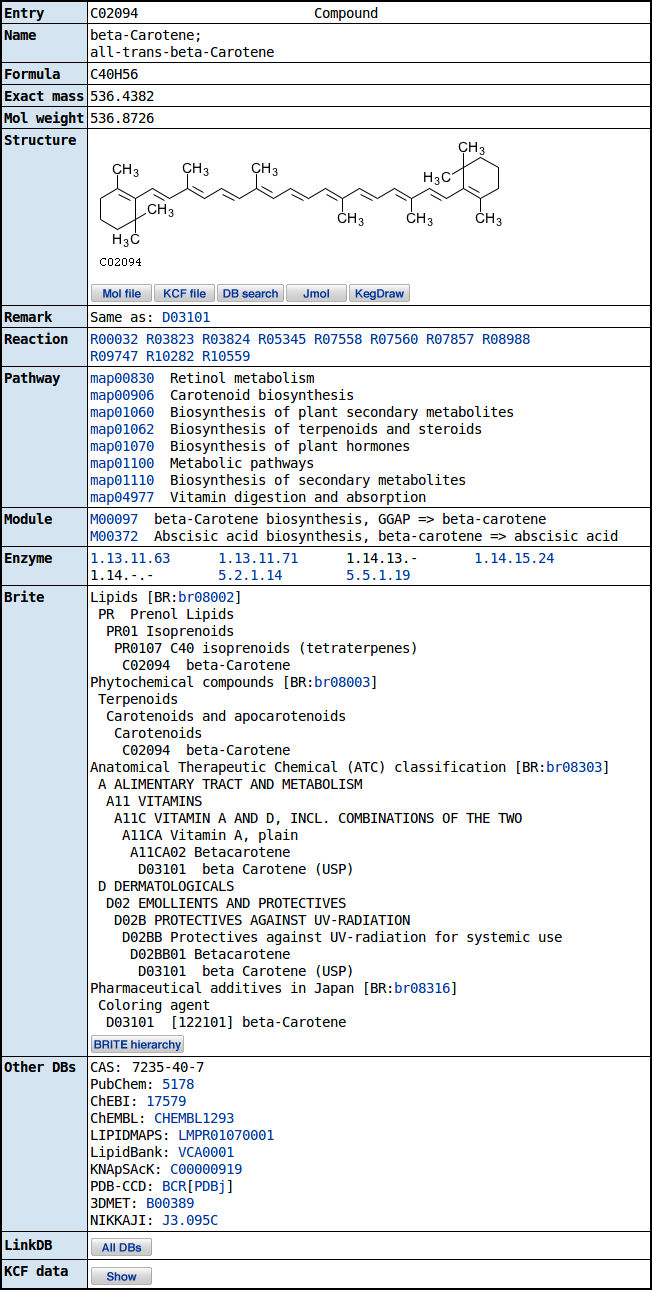
In the website you can click on the carotine biosynthesys link to see the full pathway. It looks likes
the image below. Those marked in red is bera-carotine.
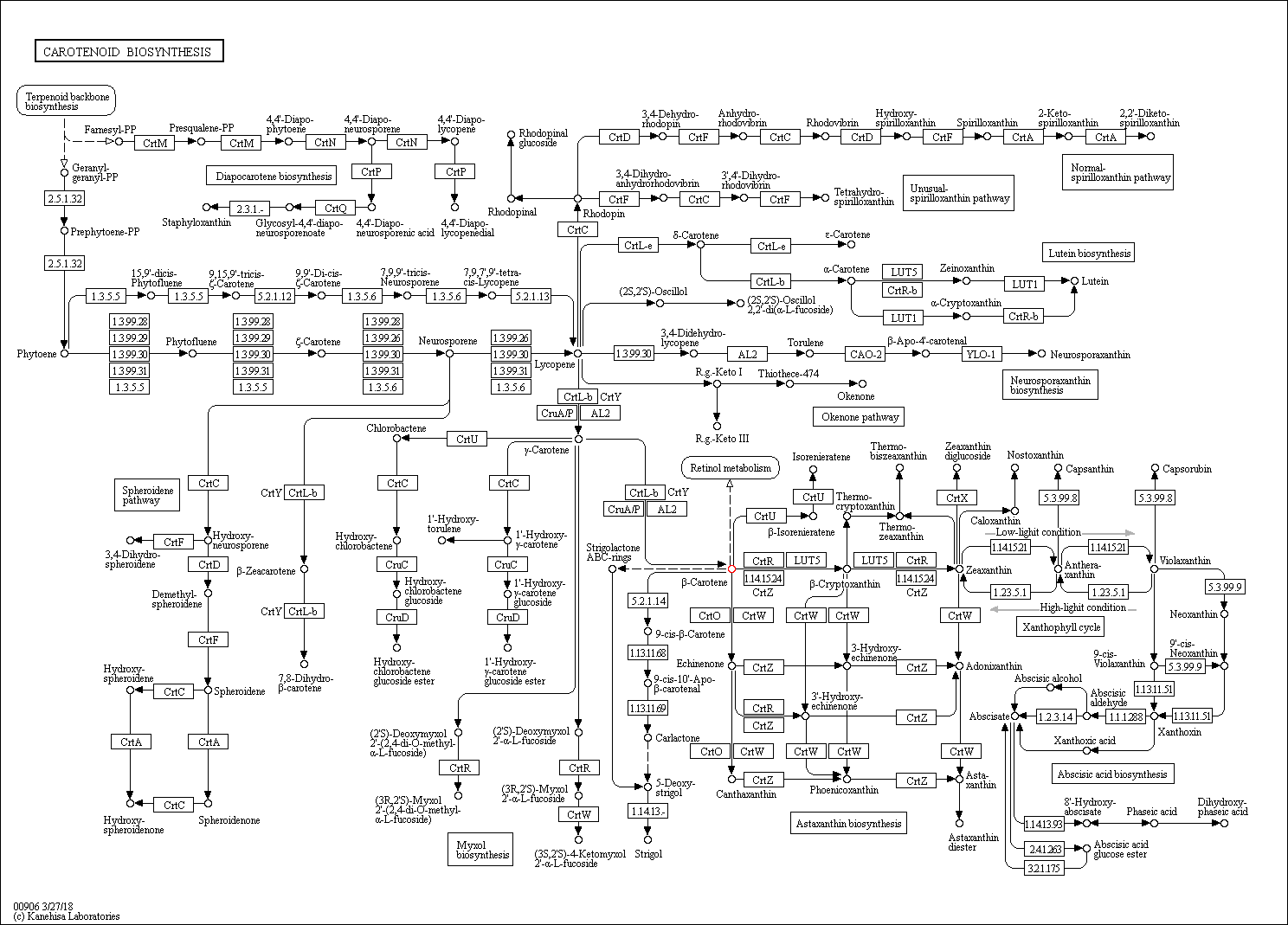
View full image
If we study the image carefully we can see that there is a pathway in which lycopene is getting converted to
beta-carotine. This is highlighted in the following image.
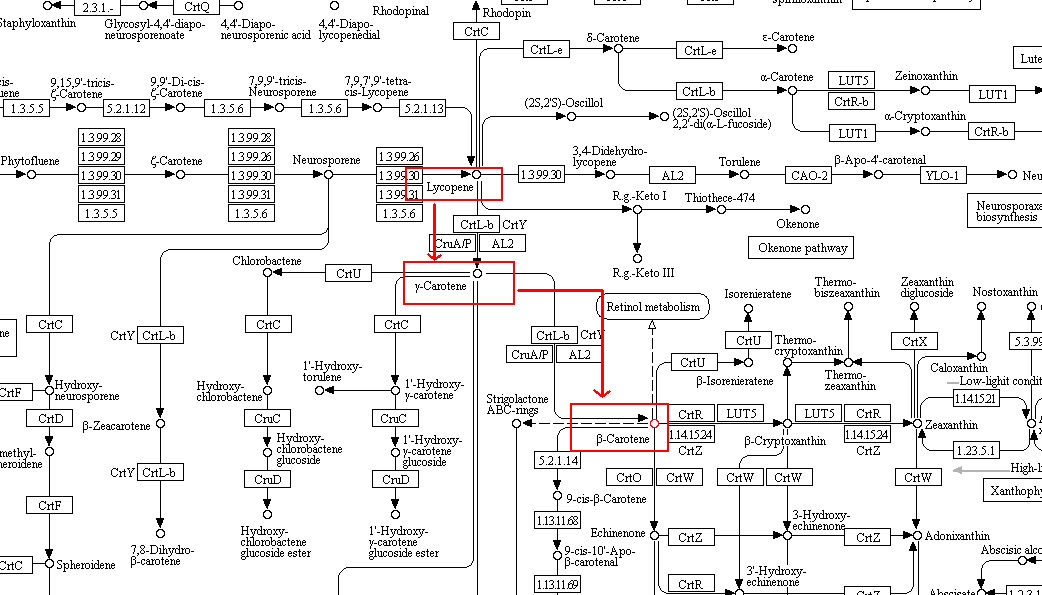
We can exploit this pathway to convert lycopene to beta-carotene. As you can see in the image, lycopene
first gets converted to gamma carotene and later gets converted to beta-carotene.
lycopene beta cyclase catalyses both the reaction.
Thus adding lycopene beta cyclase gene to lycopene producing ecoli will make it produce beta caratene instad of lycopene.
pAtLCYbSK pasmid from addgene has this particular gene. Thus transforming this
plasmid along with pAC-lyc plasmid will make an ecoli produce beta-carotene.
Ethics/ safety considerations this week
Do your activities this week raise new ethics and/or safety considerations you had not considered in week 1? Describe what activities have raised these considerations and any changes you have implemented in response.
In this week we are only transforming plasmids into e.coli. BSL1 is equipped enough to handle these kind of experiments.