Theory Task
Answer the following questions:
1. Craig Venter has stated he will establish an IGEM style competition for the most innovative use of the JCVI minimal bacterial cell, JCVI-syn3.0. The JCVI built the organism to be used as a platform to investigate the first principles of cellular life. Rather than use the cell for a basic research question, imagine that you were head of R&D at a biotech and were trying to determine if a new biochemical pathway you had developed to fix carbon from CO2 had any industrial potential. You know the method works in cell free enzymatic reactions, but that is about all you are sure of other than the enzymes and pathway. Do not get stymied by technical details involving how you would genome engineer JCVI-syn3.0. Explain the concept and experimental rationale.
Accelerated pathways to carbon fixation has a vital potential due to the all time
increase in green house gases like carbon dioxide. The greatest advantage i find in this
organism is that it is a complete living organism rathen than a synthetic minimal cell.
Thus i think it can be extensively used to test the carbon fixation pathway at different
conditions of temperature, ph etc without much interference from the host organism.
Thus for me JCVI will be providing a more flexible and versatile platform for experimentation
in a more controlled organism which is self sustainable.
2. In the previous classes, George Church has talked about work done in his lab to do grand scale genome engineering of E. coli to alter its genetic code. While there are similarities with how his lab engineers E. coli with how the JCVI engineers mycoplasmas, what is the fundamental difference or differences?
George's approach and JCVI approach are entirely different appreach for the same problem
and offers different set of merits and demerits.
Geroge's approach is a divide and conquer approach whereas JCVI approach is like a construct from scratch approach.
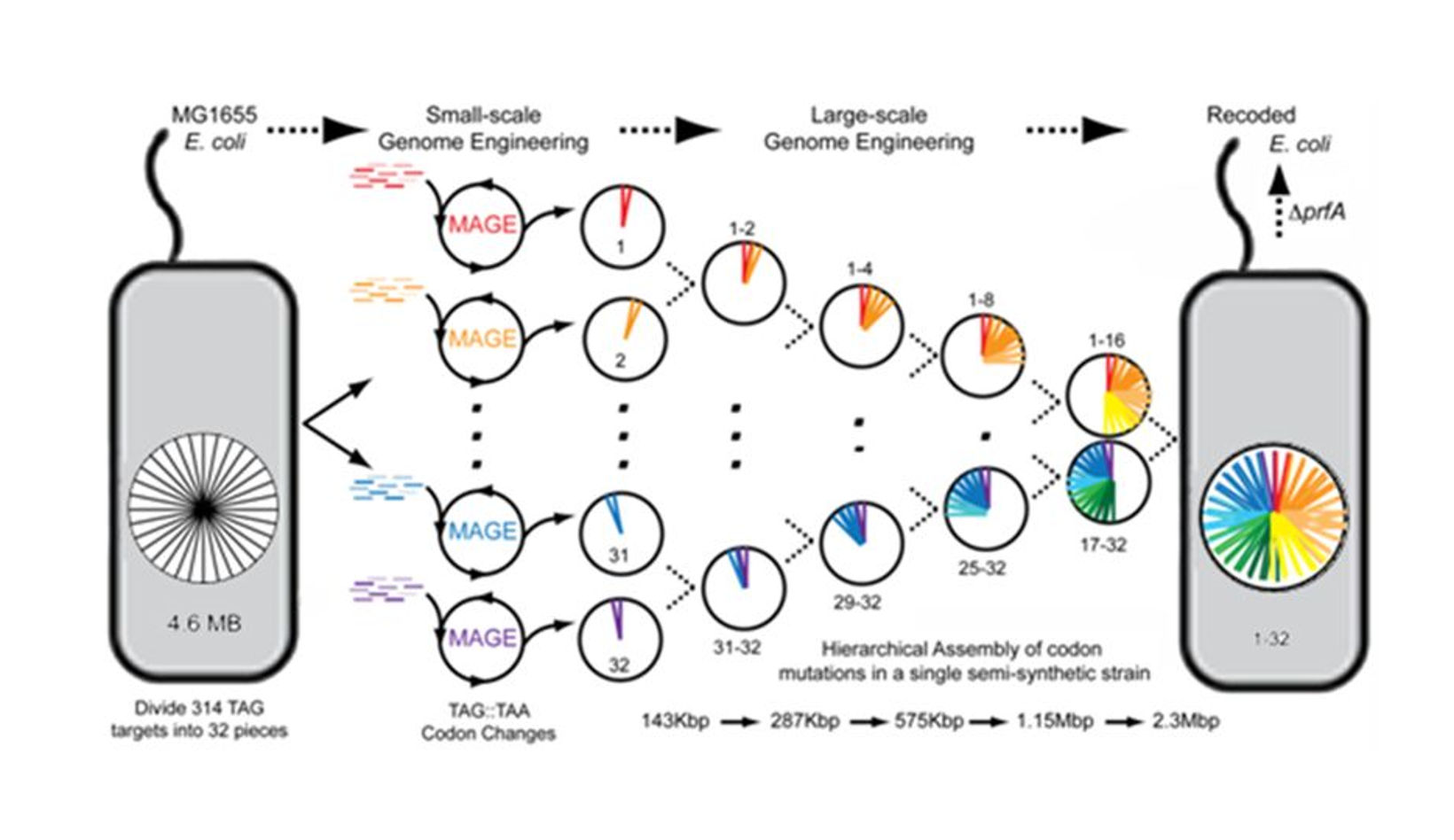
His (George's) approach is altering an existing genome using multiplex automated genome engineering (MAGE).
In this approach they will study each parts of the genome part by part and analyse its function and vitality in the
survival of that organism. The advantage of this approach is that we are not making changes all of a sudden
Whereas JCVI approach is Designing Hypothetical Minimal Genome (HMG) using transposome mutagenesis. They are creating
a new genome from scratch using existing transposome mutagenesis and deletion data.
3. In the Science paper ͞Design and synthesis of a minimal bacterial genome,͟ the JCVI showed the figure below, which is the first step in an effort to rationally reorganize the minimal cell genome. As noted in the paper, when we reorganized the genome, which included separating genes in operons, as needed we placed genes behind transcriptional promoters that controlled expression of genes deleted to build minimized segment 2. We built a genome that had a reorganized segment 2, but with the other 7 segments in with their original gene order. As noted in the paper, that cell grew normally.
In later experiments, the JCVI used the same strategy to design and build reorganized versions of the other 7 minimized segments and tested them as genomes that were 1/8th reorganized and 7/8ths not reorganized. None of those 7 genomes resulted in viable cells. What is your hypothesis as to why this happened?
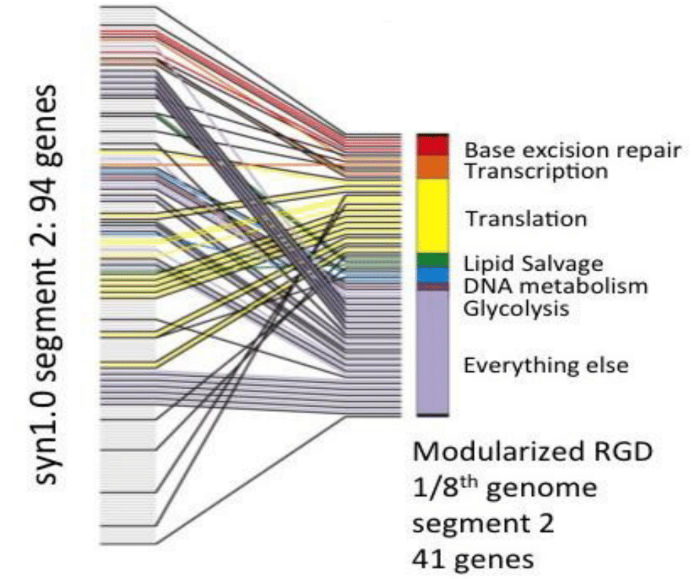
As per my understanding the problem might be dividing the genes in operons and dividing
them based on the process in which they are involved. The later worked and the former
failed.
4. The JCVI recently announced that it was now minimizing and reorganizing the genome of the fastest growing eukaryote, a yeast called Kluyveromyces marxianus. The goal will be to make an alternative to Saccharomyces cerevisiae that grows faster, and can be grown at higher temperature, and that would be a better platform for the kinds of biotechnology people currently use yeast for. Based on the ͞genome engineering lecture in HTGAA and the Science paper ͞Design and synthesis of a minimal bacterial genome͟(assigned as class reading), how would you go about minimizing and rationally reorganizing the K. marxianus? What would be different about your approach relative to how the JCVI minimized Mycoplasma mycoides to produce JCVI-syn3.0?
Kluyveromyces marxianus is a eukaryote and its genome is much more complex than ecoli, also the presence of non coding regions called introns will add to the complexity.
Ethics/ safety considerations this week
Do your activities this week raise new ethics and/or safety considerations you had not considered in week 1? Describe what activities have raised these considerations and any changes you have implemented in response.
There are no ethical/safety concern for this week.