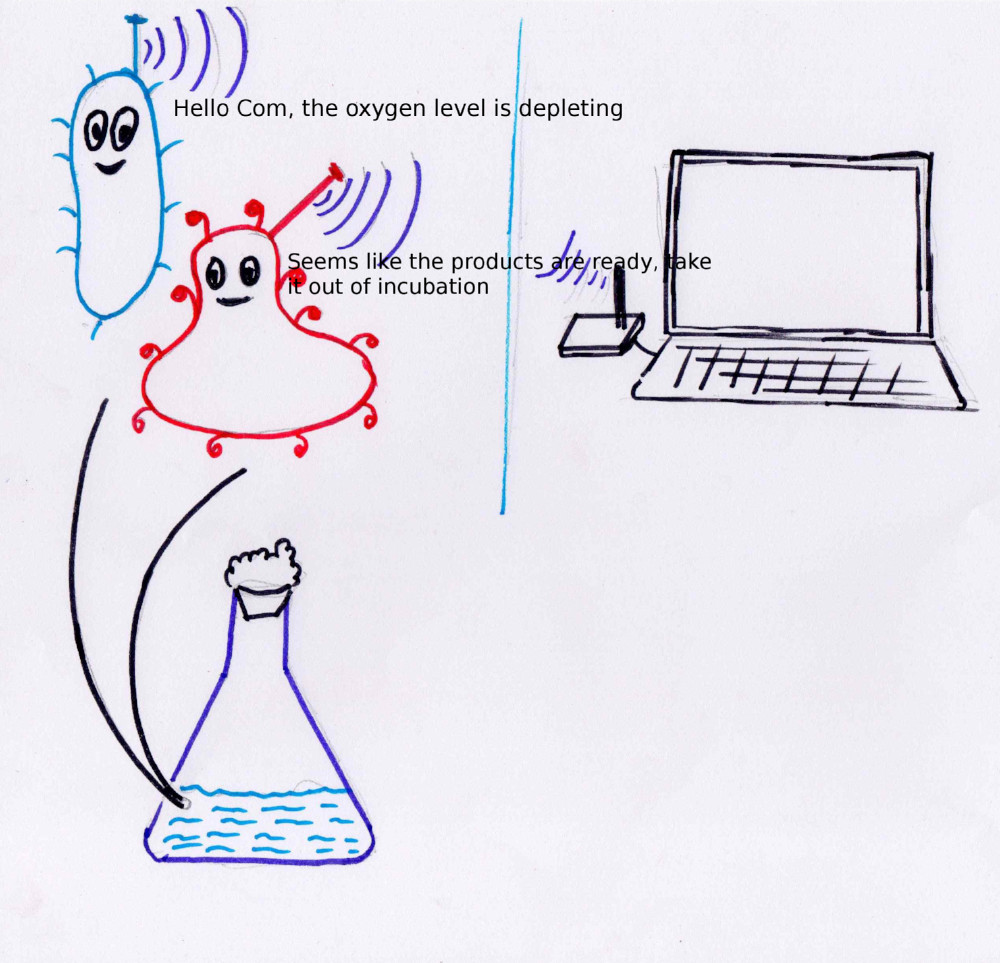
I have this dream of a day when we will be sending out micro organisms into the space which invade into the darkest corners of the universe and send us information about what they see and sense in those remote planets and satellites, the day when friendly microbs living in our body will report us real-time on the functioning of the body and the possibility of a potetial diseases. This course is a step forward towards my dream.
I believe that synthetic biology and allied technology are fast approaching in that direction of mastering the understanding of life which will give us the power to manipulate life forms in the way we want. One major necessity which i think will super charge this field would be a spontaneous communication mechanism from and to the micro organism. In this project i want to take a different direction in manipulating life forms in a way they can communicate directly with us in realtime and possibility extent it to a process wherein we will be able to directly communicate with the micro organism and ask it to transcribe a protein of interest.
Majorly studied and understood communication pathways in micro organism are purely
chemical communications pathways and if they
are unicellular organism them the communication is mostly internal. Usually this
type of communication happens to convey the
presence or absence of specific molecules in the surrounding. Synthetic biology has
exploited this by artificially controlling
such molecules in the surrounding to the make the cell do something. For example
today we can control the presence of glucose and
program a cell machinery to specifically look for glucose concentration and express
a particular gene in the presence of this
molecule.
On the other side our electronics and computational industry have progressed
tremendously and so many things which was previously
impossible to do are possible to do
today.There is a huge potential of could connect life science with electronics and
communication technology.
The problem with connecting the synthetic cells are as a sensory of actuator unit
with our electronic technology is
the communication issue. Even if could make a living cell based bio sensor it would
be hard for it to communicate with our computers
as they speek the language of chemicals and computers speak the language of 1s and
0s.
This project is a proposal for bridging that communication gap between our present
day electronics technology and the synthetic biology.
In simple words i am exploring a method by which micro organisms can communicate
with our electronic devices and we can communicate with the
cells, asking them to do something using a physical signal.
Starting the search
I started searching for a instance wherein a micro organism generates
electromagnetic radiation preferably of the
RF range which can directly be used as a communication tool between the organism and
our electronic devices.
But search didnt take me anywhere except for this paper which tried to prove that
bacteria in
bio films use EM signals to recurit other microbs. But it was not well defined on
how they do that nor how we can
reproduce that in the way we want. It is then that i realized that there is already
a well studies, well documented
instance in the synthetic world wherein a bacteria produces electromagnetic signals
in 490 nanometers.
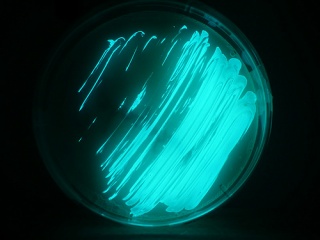
Yes i am
talking about biolumilance by Vibrio fischeri and Vibrio harveyi. These bacteria has
genes coding for luciferece and Luciferin
which on reacting produces light. This is pretty good as our systems are evolved
enough to detect, record amplify and transmit light
as a signal. Also optical fiber communication takes place with light signals. Thus
bioluminance is that interface between that
chemical signals which live cells speak and physical signals which our electronic
devices communicate with.
Bacterial bioluminance
Biochemistry of the Bacterial Bioluminescence
Bacterial luciferase and the enzymes that supply and regenerate the substrates of
bacterial
luciferase are the key components on
bacterial bioluminescence. The catalytic machinery involved in continuous
light production in luminous bacteria is the enzymatic reaction between luciferase
and luciferin
The DNA sequences coding the proteins in the luminescent system are popularly known
as lux
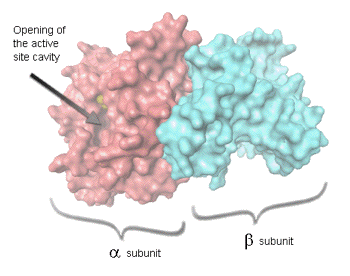
genes. Bacterial luciferase is a heterodimer, composed of two different
polypeptides, called alpha and beta (of molecular mass 40 kDa and 37 kDa,
respectively, and encoded by the luxA and luxB genes, respectively. The active site
is located within the subunit.
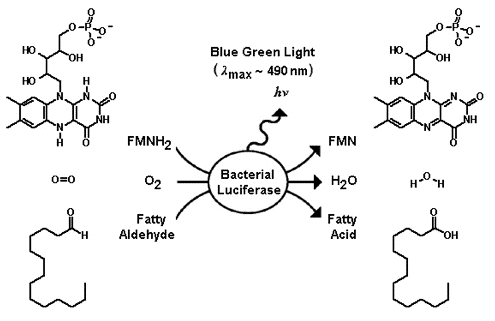
The substrates of bacterial luciferase are reduced flavin mononucleotide (FMNH2),
molecular oxygen, and long chain fatty aldehyde. The excess energy, which
is liberated from the oxidation of FMNH2 and aldehyde concomitant with the reduction
of molecular oxygen, is released as blue/green light emission (~ 490 nm).
The characteristic color indicates the energy level of the photon that was produced
when the excited electron on the flavin chromophore returns to the ground state.
Researchers have found that flavin analogs with substituted atoms
in the chromophore moiety resulted in different luciferase emission colors.
Tt has also been shown that point mutations at the flavin chromophore's
binding site distorts the color emission spectrum of bacterial bioluminescence,
indicating that the distinctive emission color depends not only on the chromophore
that emits the photon, but also the electronic nature of the chromophore-binding
microenvironment in luciferase. Aside from bacterial luciferase, some luminous
bacteria carry fluorescent proteins to modulate the emission color, distinguishing
themselves from other strains. Interaction of blue fluorescent protein (also known
as the lumazine protein) in Photobacterium phosphoreum and Photobacterium leiognathi
with the respective luciferases yields photon emission at higher energy ( ~478
nm) corresponding to a blue color. Similarly, the presence of the yellow
fluorescence protein in the strain Y-1 of V. fischeri results in yellow light
emission ( ~545 nm). The participation of the fluorescence proteins in the
lucifrease-catalyzed reaction also alters the reaction kinetics of bacterial
luciferase.
In order for light emission to occur for long period of
times, substrates must be supplied continuously to bacterial luciferase. As the time
frame and the level of product generation in enzymatic reactions are limited by the
availability of the substrates, the constant light emission in luminous bacteria
must therefore be maintained by several different enzymes continuously generating
the substrates for the bioluminescence reaction. Those enzymes that replenish the
aldehyde substrate are coded on the lux operon; in particular, the fatty acid
reductase, a multienzyme complex, whose lux genes (luxC, luxD, and luxE) immediately
flank the luxA and luxB genes of luciferase.
A critical component in the biochemistry of bacterial bioluminescence is molecular
oxygen, which is supplied from the external cellular environment. Without the input
of molecular oxygen, luminous bacteria cannot emit light. In the bacterial
luciferase-catalyzed reaction, the energy expenditure on the reduction of molecular
oxygen to a peroxy reaction intermediate, and then ultimately to water serves as a
trigger for releasing the potential energy from the oxidation of both FMNH2 and
fatty aldehyde in the form of photon emission.
Mutating other bacterias for bioluminance
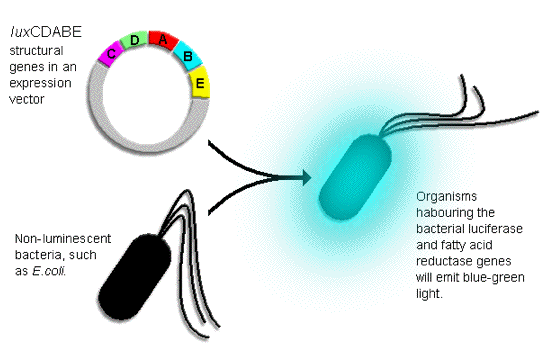
The lux operon gene is responsible for bioluminance in bacterias. This gene can be
transferred to other bacterias including
ecoli to make them emit light. The gene which codes for the substrate and the enzyme
togather are called luxCDABE genes which
can be mutated into other bacteria to make them bioluminacent.
The project definitions
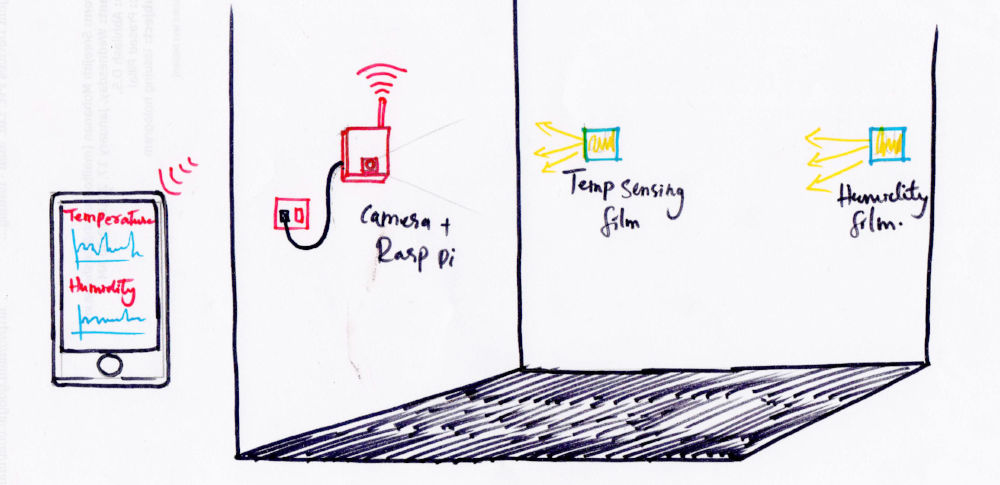
Due to the constrain in time and resources and present-ability i am boxing the
project into an IOT based
environment monitoring system wherein genetically mutated ecoli cells with induced
bio luminance will be used as sensor to
monitor environmental factors like temperature, humidity etc and report directly
with the intensity of expressed luminance. Thus if it is a temperature sensing
culture, the luminance produced by
the culture will be proportional to
the intensity of light emitted my them. Same with humidity and other factors.
Using genetically modified bacteria for sense temperature and
humidity maight look silly as we have cheap hardware for that, but this kind of
sensors has the potential when if comes to specific
cantaminant/pathogen/allergen sensing eg: lead and copper quantity measurement
wherein traditional sensors fail.
On the other side of the room the luminance reported by the culture will be
continoulsy observed by a camera mounted computer. Here in
this project i am using Raspberry pi 3 and piCam and build in wifi to do that. I
will use opencv to first identify the position of
luminance in the whole image and then use localized color averaging to find a single
value representation for the
observed intensity. This value will be studied against actual temperature
measurement, developing a co-relation
between the measured values of color intensity and the actual temperature. A single
camera will be able to get data from multiple such
sensors.
Now this data can be synced directly into a cloud server and can be directly view on
a monitor or a desktop computer.
The project accomplishment is that we use independent minicultures of GM ecoli as sensors and reportes for an electronic device. In a way we are interfacing live cells as a part of electronic device.
The Hardware and software
Here as mentioned i am using Raspberry pi 3 as my soc and Picam which is a camera
specifically build for the raspberrypi as the camera
for my setup. Setting up raspberry pi is quite easy. You just have to download the
OS image from the website and burn it into a micro sd card.
This will make the sd-card bootable. You can then simply put this card into the pi
and connect the monitor, keyboard etc and turn on the power.
You can see the computer booting up with a nice looking GUI.
For this purticular project we want our camera to be continously looking at a
luminascent ecoli culture and simultaniously read the
intensity value. For this purpose we need to use any computer vision library. The
popular and open sourced one out there is
opencv which has python binding which is quite easy to use. The major pain is
installing opencv in the PI. none of the python package managers
can help you with that. You will have to download the source and build it by
yourselves. And often you will find so many errors on the way.
If you could successfully install opencv in the system the job is half done.
The culture plate
Next part of the work is designing and making the culture plate. I made a quick
drawing of the extimate structure
of the plate.
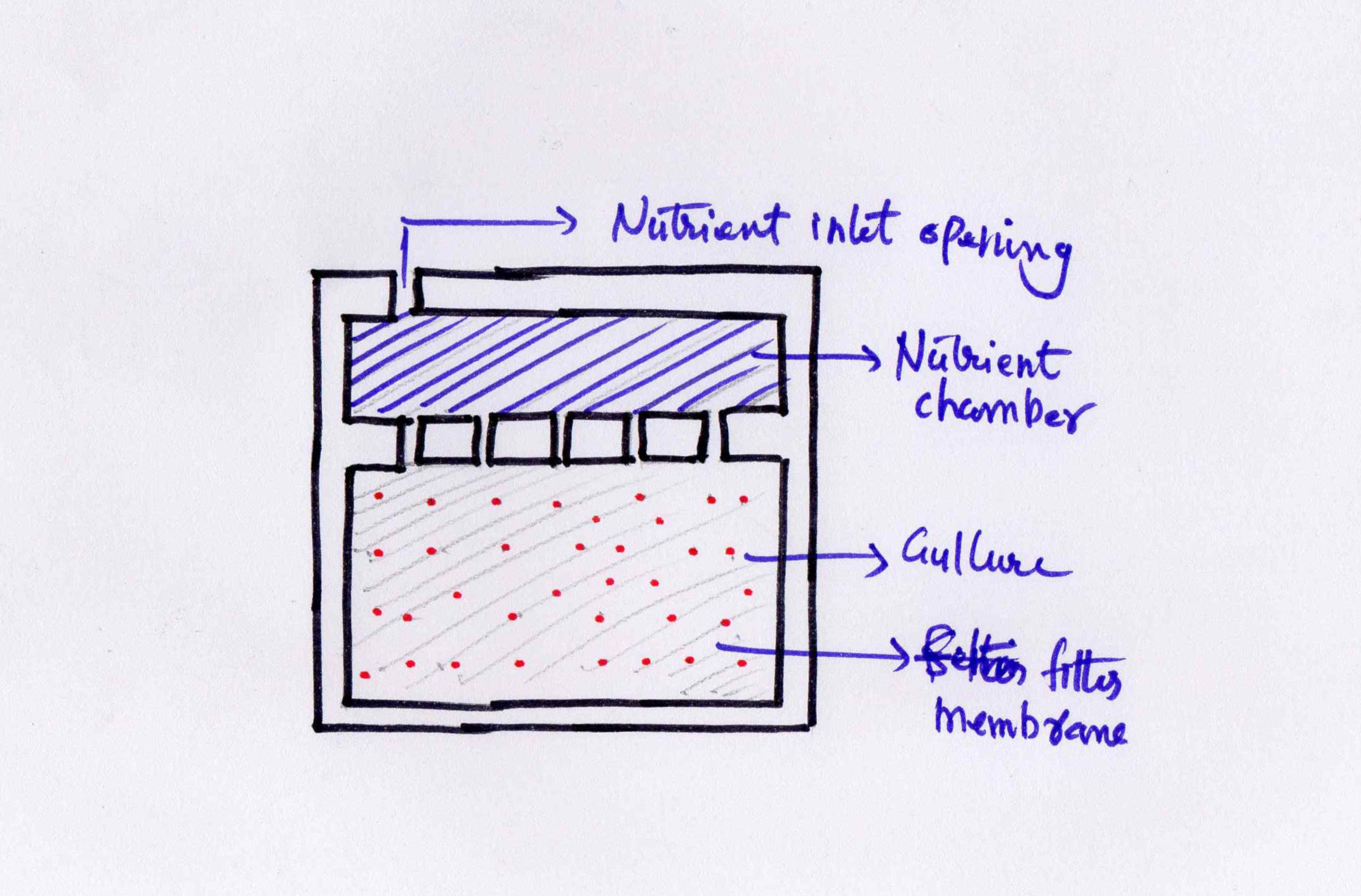
This apparatus i am planning to cut in laser. It has two chambers. THe top chamber
is for supplying nutrients to the medium
containing the bacteria in the bottom chamber.
Final demo
Limitations
- In the light the system is not properly visible to the camera, thus the system shows good result only in the dark
- Sometimes other environmental factor maight affect the luminance of the bacteria causing wrong results