Next Generation Synthesis
Theory Assignment
Design a gene. Describe a detailed workflow for constructing and expressing it. Identify how the parts of your genetic construct relate to DNA replication and the Central Dogma of Molecular Biology.
The parts of a gene and their functions are as follows from
IGEM Parts Registry website:
- Promoters: A promoter is a DNA sequence that tends to recruit transcriptional machinery and lead to transcription of the downstream DNA sequence.
- Ribosome Binding Sites: A ribosome binding site (RBS) is an RNA sequence found in mRNA to which ribosomes can bind and initiate translation.
- Protein coding sequences: Protein coding sequences encode the amino acid sequence of a particular protein. Note that some protein coding sequences only encode a protein domain or half a protein.
- Terminators: A terminator is an RNA sequence that usually occurs at the end of a gene or operon mRNA and causes transcription to stop.
Workflow for constructing and expressing a gene
The gene of interest is the gene that is responsible for blue eyes in Homosapiens. Mutations in the human melanosomal P gene were responsible for classic phenotype of oculocutaneous albinism type 2 (OCA2). The P protein isoform 1 was taken from the NCBI website. for constructing the gene and hence express it [1]
- The primer was designed from Primer 3 website
- The DNA sequence of P protein isoform was pasted and the OCA2 gene was selected for the selection of primers with the default parameters.
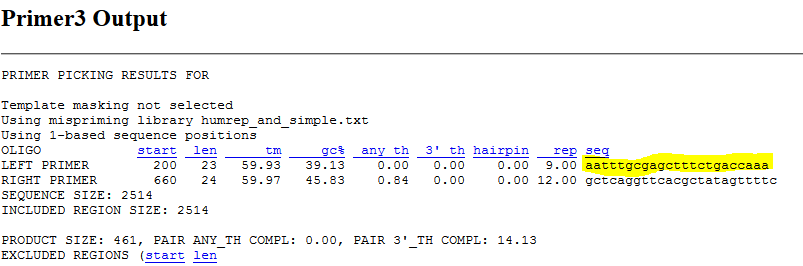
-
The forward primer with an extension of the 5' end with random sequences was selected and the reverse primer in the 3' strand (bottom strand selected). The screenshot of selection of forward and reverse primers in Snapgene as shown below:

- Amplification of the OCA2 gene between 338 bp and 838 bp amino acid sequence i.e.(1014 to 2514 bp nucleotide sequence) is then done for the expression of blue coloured eyes, according to NCBI website
- Amplification is done using PCR in Snapgene from PCR option in Actions Tab. The screen of selection of primers for PCR is as shown in the image below
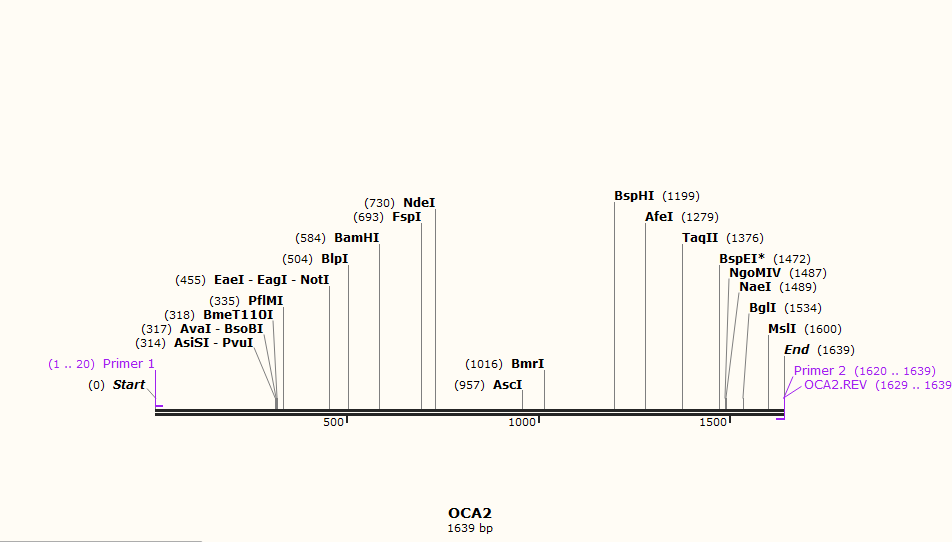
- The protein is then made from Tools tab - Make a protein option (translation) after the amplification of the sequence using PCR.
- One or more open reading frames or codons are to be selected for making the protein. The ORF between 1014 to 2514 bp is selected for the purpose of OCA2 gene.
The translated protein of the selected sequence is shown in the below image which corresponds to the OCA2 gene.
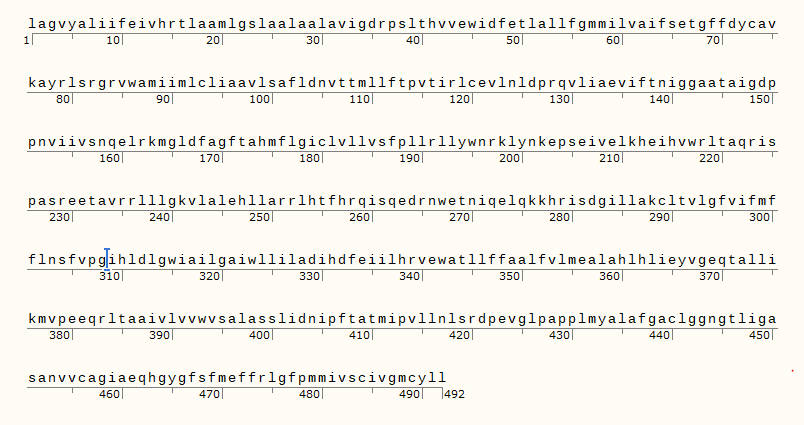
Relation of the part of genetic construct to Central Dogma of Molecular Biology
The below schematic diagram shows the Central Dogma of molecular biology:
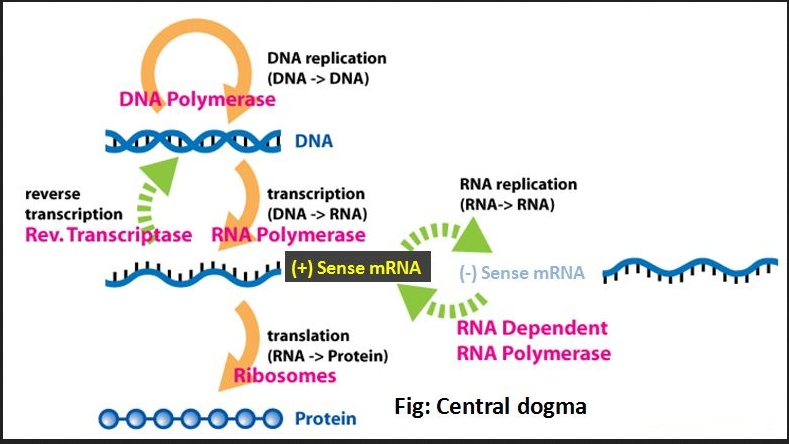
- The primers (a short strand for initiating the DNA synthesis) are first selected. It is required for the DNA replication, the first step in Central Dogma, where the DNA polymerase only helps in adding new nucleotides to the existing strand in the the 3' end.
- Later the amplification of the selected DNA sequence is done using PCR. The making of protein from the existing DNA sequence called translation is done, which expresses the OCA2 gene.
Practical Assignment
Recode green fluorescent protein (GFP) to instead emit blue or cerulean light. Confirm success by transforming your assembled BFP and CFP variants into E coli cells.
Cloning experiment will be started once the oligos arrive (delay in shipping) and the experiment and the results will be updated after that.
Do your activities this week raise new ethics and/or safety considerations you had not considered in week 1? Describe what activities have raised these considerations and any changes you have implemented in response.
- The design of the gene using software did not raise new ethics or safety considerations. But from the practical viewpoint, I understood that the mutations should be very carefully done for the betterment of the people on earth, else the earth might turn dangerous for life
REFERENCES
[1] Eiberg, H., Troelsen, J., Nielsen, M., Mikkelsen, A., Mengel-From, J. and Kjaer, K.W., 2008. Blue eye color in humans may be caused by a perfectly associated founder. Hum. Genet, 123(2), pp.177-187.
Go back to home page

