Biodesign
Design Assignment
Choose a DNA sequence that interests you from nature or a source like the iGEM part registry. Provide a detailed workflow for isolating or acquiring a molecule with this specific sequence and an assay that confirms part of its content.
Detailed Workflow for Isolating a DNA molecule
Isolating DNA using Restriction Enzyme Digestion:
Since childhood, the emission of light from fireflies has inspired me. The enzyme luciferase makes the firefly emit light and it has inspired me to isolate the DNA sequence that is responsible for the bioluminescence.
A short description about pARE- lux plasmid:
pARE-lux plasmid is consisting of the vector backbone pGL2- Basic, the method of cloning of the plasmid is by Restriction enzyme. It is resistant to Ampicillin and the growth of the plasmid is at 37 C and quite fast. The plasmid consists of three copies of the ARE (activin response element) from the Xenopus Mix.2 promoter linked to a basic TATA box and a luciferase reporter gene[1].
Protocol
Assay for confirming the part of its content
Gel electrophoresis is the method of visualizing if the digestion has taken place properly. Gel electrophoresis is the standard lab procedure for separating DNA by size (e.g., length in base pairs) for visualization and purification. Electrophoresis uses an electrical field to move the negatively charged DNA through an agarose gel matrix toward a positive electrode. Shorter DNA fragments migrate through the gel more quickly than longer ones. Thus, you can determine the approximate length of a DNA fragment by running it on an agarose gel alongside a DNA ladder (a collection of DNA fragments of known lengths).
Verifying the restriction digestion:
The purpose of choosing 2 restriction enzymes for cutting the same sequence at the same site, is because we would be confident about the restriction digestion process. The agarose gel is prepared, comb is inserted and gel is solidified. Ethidium bromide is added, which is a known mutagen, that binds to the DNA, and the fragments of the DNA can be visualized in the presence of UV light.The samples are loaded on the wells in the gel and 80 to 150 V supply is given in order to run the gel. Then the gel is visualized using any instrument that possess UV light. This assay confirms the process of restriction digestion.
Practical Assignment
Plasmid Map
- Download Snapgene from the website
- Opened a new file and inserted the pUC19 plasmid sequence got from Addgene
- Plasmid map appeared on the map tab as below:

- Searched for the PvuII restriction enzyme and found that the enzyme cuts twice at 54 and 376 bp in the lacZalpha.

- Simulated the gel electrophoresis in Snapgene in Tools tab in order to cross check with the practical experiment. The below image shows the simulated agarose gel.

Practical Assignment
Verify restriction digest of a circular DNA plasmid by gel electrophoresis. Compare your experimental result with what you would predict from the plasmid map.
- I have taken the reagents pUC vector, PvuII enzyme, Cutsmart Buffer 10X and deionized water at the concentrations mentioned below.
- pUC vector - 5muL
- PvuII - 2.5muL
- Cutsmart Buffer - 7.5muL
- Deionized water - 60muL

- The samples were separated to 30uL each and one was stored at 37C and the other at 4C.
- Meanwhile, 1% agarose gel was prepared. 0.5g agarose in 50ml water was measured, microwaved for 1 minute 30 seconds till the agarose is dissolved.
- The tray was inserted with a comb and once the agarose solution is cooled, 5ul of Nancy-520 was added to the agarose solution. The agarose solution was then poured into the tray and waited for the gel to solidify for about half an hour.
- Then the samples were added into the wells in the gel in the following order - DNA Ladder, Negative control with the uncut plasmid, one sample of PvuII digested at 37C and other sample of pUC19 plasmid cut with PvuII at 4C.
- 1X TAE Buffer of about 1000ml was prepared from the existing 50X TAE Buffer. Approximately 750ml of TAE buffer was added till the gel is immersed and the gel electrophoresis apparatus was set at 120V and was run.
- The gel was then run 3/4th the length of the gel and observed with Gel Imaging Apparatus for the restriction digestion of pUC vector with PvuII enzyme.
- The gel electrophoresis apparatus in the BioNest facility is as shown below.

- It appeared that the gel was not run properly and didnt have well defined bands even for the 1kb DNA Ladder. This might probably be due to the fact that we prepared the gel previous day and was kept inside the fridge wrapped in an aluminium foil till the next day, may be the wells were not well cut, or probably might be due to an improper current flow in the apparatus, since the electrodes were not well tightened. The below image shows the image of the gel.

- So I repeated the experiment by preparing a fresh 1% agarose gel, crosschecked that the anode and cathode connected are intact and run it again with the same set of samples for 1 hour.
- To crosscheck if the gel is running properly, we paused in between after 30 min and checked with LED Transilluminator if there are good well defined bands. The image is as shown below

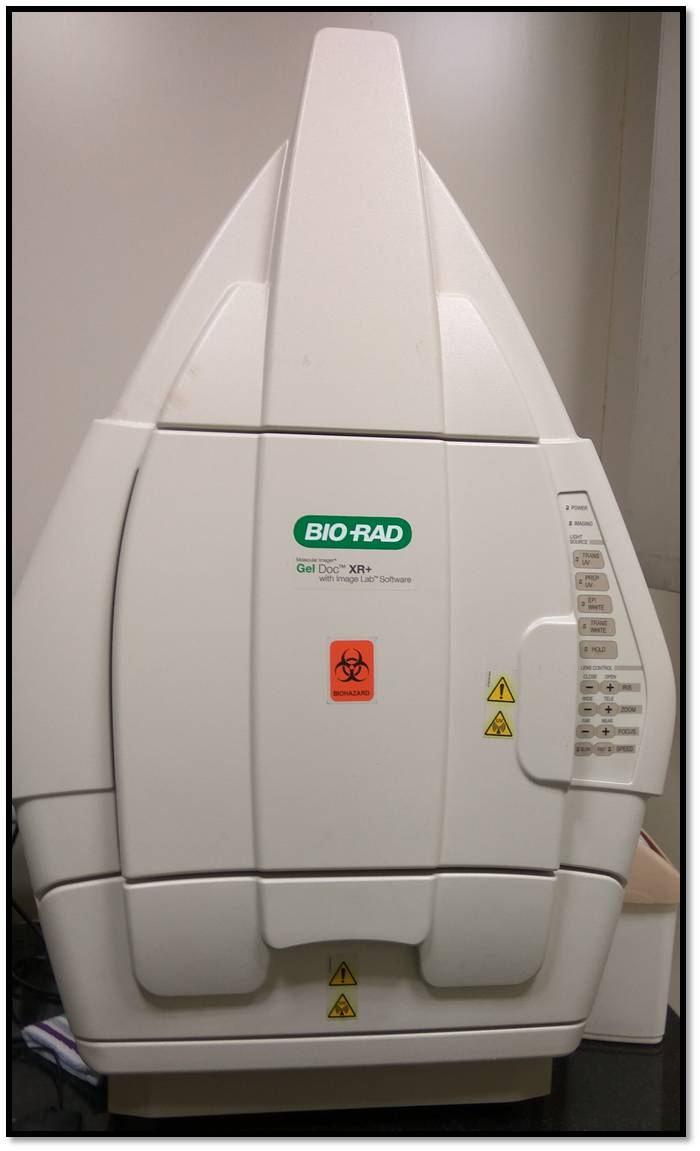
- The gel was then imaged using the BIORAD Gel Doc with Image Lab Software available at BioNest. The image of the apparatus is shown below:
- The below image shows the gel run for the second time.

- When the second gel image was compared with that of the simulated gel from Snapgene, I find that both the images look alike and the restriction digestion has happened in both 37C and 4C.
New ethics or safety considerations this week
Do your activities this week raise new ethics and/or safety considerations you had not considered in week 1? Describe what activities have raised these considerations and any changes you have implemented in response.
- The stain that we used is Nancy-520. The stain was safe to use and stain did not raise any safety considerations.
- The laminar flow was on along with UV, and got to know that UV should be separately switched off when working in the cabinet. UV burns the skin if working with UV on in the cabinet.
REFERENCES
[1] TGF beta signals through a heteromeric protein kinase receptor complex. Wrana JL, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massague J. Cell. 1992 Dec 11. 71(6):1003-14. 10.1016/0092-8674(92)90395-S PubMed 1333888
Go back to home page





