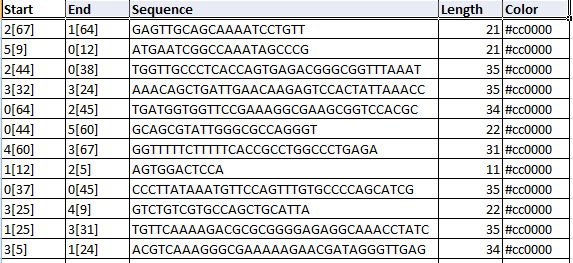
The below image shows the sequences along with the colour code in ASCII format and the length of the sequence.
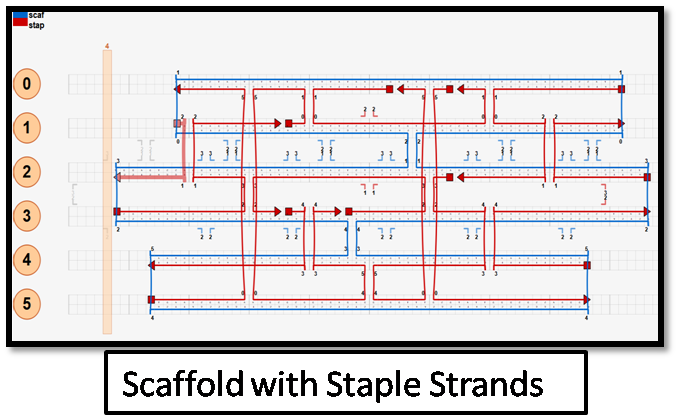
I installed caDNAno, Autodesk Maya and Python in Windows. With caDNAno, I created a M13 scaffold and then created staple strands and used AutoBreak Option to create strands inside the scaffold.
I learnt the basics of editing in cadnano, to use tools like pencil, select, add sequences, auto staple, autobreak, etc.
With the help of the tutorials available, I just tried out. The screenshots are as shown below:
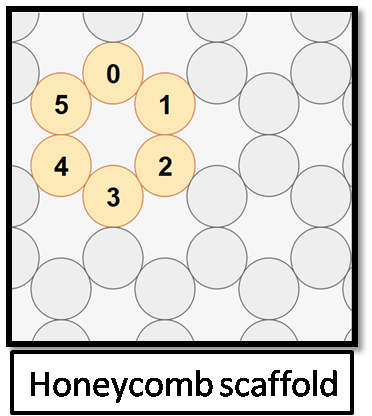
The scaffold I created is in blue lines and the staple strands indicated in red (2nd picture) from the honeycomb structure (1st picture), as given below:


The below image shows the sequences along with the colour code in ASCII format and the length of the sequence.

With the above understanding, I am going to create nanostructures for an interesting application.
For the DNA Paint experiment, we have got 3 eppendorf tubes labelled P0, P1 and P2. From my understanding, the P0 is the oligos and P1 and P2 are HCR initiators.
Problems faced:We have the facility to do PCR. But I was not given permission to keep the thermocycler switched on for 173 hours as there are many researchers using the PCR on a regular basis.
Also I got to know that only if I am successful in Expansion Microscopy, I will be able to do the DNA PAINT experiment. I did not get a clarity on the same. When approached Teja from Cambridge for help, he had suggested me an alternate protocol for the same without PCR. So I had tried it out with the DNA oligos that was shipped.

We observed that the oligos did not get crosslinked with the protocol that we followed.We intend to repeat the protocol with PCR again and see how it goes.
For the TEM analysis,I intend to enquire at RGCB in Trivandrum or AIMS Facility at Cochin to visualize the interlinked oligos.